The bioinformatics facility at NCCS provides access to high-performance compute resources and programming expertise. The compute infrastructure serves scientists at NCCS to master the informatics needs of their research in a proficient and cost-effective manner.
- Databases
- Docking Tools
- List of PPI Servers
- Network Analysis Tools
- Signaling and Metabolic Databases
- System Biology Tools

Databases
|
Sno |
Database |
Site |
Use |
|
Protein |
|||
|
|
Uniprot |
Protein knowledgebase database consists of two sections: (A) Swiss-Prot: manually annotated and reviewed. (B) TrEMBL: automatically annotated and is not reviewed. Includes complete and reference proteome sets. |
|
|
|
UniRef |
Sequence clusters, used to speed up sequence similarity searches. |
|
|
|
UniParc |
Sequence archive, used to keep track of sequences and their identifiers. |
|
|
|
NCBI |
Protein database collects sequences from several sources, including translations from annotated coding regions in GenBank, RefSeq and TPA, as well as records from SwissProt, PIR, PRF, and PDB. |
|
|
|
PRINTS |
A collection of protein fingerprints (conserved motifs used to characterise a protein family) |
|
|
|
PIR |
Protein Information Resource (PIR), an integrated public bioinformatics resource to support genomic, proteomic and systems biology research and scientific studies |
|
|
|
Pfam |
The Pfam database is a large collection of protein families, each represented by multiple sequence alignments and hidden Markov models (HMMs) |
|
|
Genes |
|||
|
Genebank |
Text and similarity searching of the GenBank sequence database provided by the National Center for Biotechnology Information (NCBI). |
||
|
Genedb |
GeneDB is a genome database for eukaryotic and prokaryotic pathogens |
||
|
NCBI |
Gene integrates information from a wide range of species. A record may include nomenclature, Reference Sequences (RefSeqs), maps, pathways, variations, phenotypes, and links to genome-, phenotype-, and locus-specific resources worldwide. |
||
|
Structure |
|||
|
1. |
RCSB (The Research Collaboratory for Structural Bioinformatics) |
The Protein Data Bank is a repository for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. |
|
|
2. |
CSD ( Cambridge Structural Database) |
The repository of small molecule crystal structures |
|
|
3. |
ICSD ( Inorganic Crystal Structure Database) |
ICSD is a database of inorganic crystal structure data, contains information on inorganic crystal structures published since 1913, including pure elements, minerals, metals, and intermetallic compounds (with atomic coordinates). |
|
|
4. |
BTPRED (Beta-Turn Prediction Server) |
BTPRED predicts the location and type of beta-turns in protein sequences. Predictions are made using a combination of artificial neural networks and simple filtering rules. |
|
|
5. |
CATH / Gene3D |
CATH classifies protein structures (downloaded from the Protein Data Bank) and domains into superfamilies’ when there is sufficient evidence that they have diverged from a common ancestor. |
|
|
6. |
Swiss model repository |
It is a repository for protein structure homology models |
|
|
Phylogenetic analysis |
|||
|
1. |
PHYLIP ( PHYLogeny Inference Package) |
PHYLIP is a free package of programs for inferring phylogenies. |
|
|
2. |
Phylogeny.fr |
Phylogeny.fr web server is dedicated to reconstructing and analyzing phylogenetic relationships between molecular sequences. |
|
|
3. |
moyble@pasteur |
Create workflows and save them for fast and easy reuse |
|
|
Alignment |
|||
|
(A) |
Global alignment |
||
|
1. |
CLUSTALw |
||
|
2. |
Clustal omega |
Clustal Omega is a new multiple sequence alignment program that uses seeded guide trees and HMM profile-profile techniques to generate alignments. |
|
|
3. |
Clustal w2 |
ClustalW2 is a general purpose multiple sequence alignment program for DNA or proteins. |
|
|
4. |
Expasy |
||
|
5. |
MAFT |
Multiple alignment program for amino acid or nucleotide sequences |
|
|
(B) |
Local alignment |
||
|
1. |
blastp |
http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE |
Search protein database using a protein query Algorithms used: blastp, psi-blast, phi-blast, delta-blast |
|
2. |
blastn |
http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE |
Search a nucleotide database using a nucleotide query Algorithms used: blastn, megablast, discontiguous megablast |
|
3. |
blastx |
http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE |
Search protein database using a translated nucleotide query |
|
4. |
tblastn |
http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=tblastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC |
Search translated nucleotide database using a protein query |
|
5. |
tbblastx |
http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=tblastx&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC |
Search translated nucleotide database using a translated nucleotide query |
|
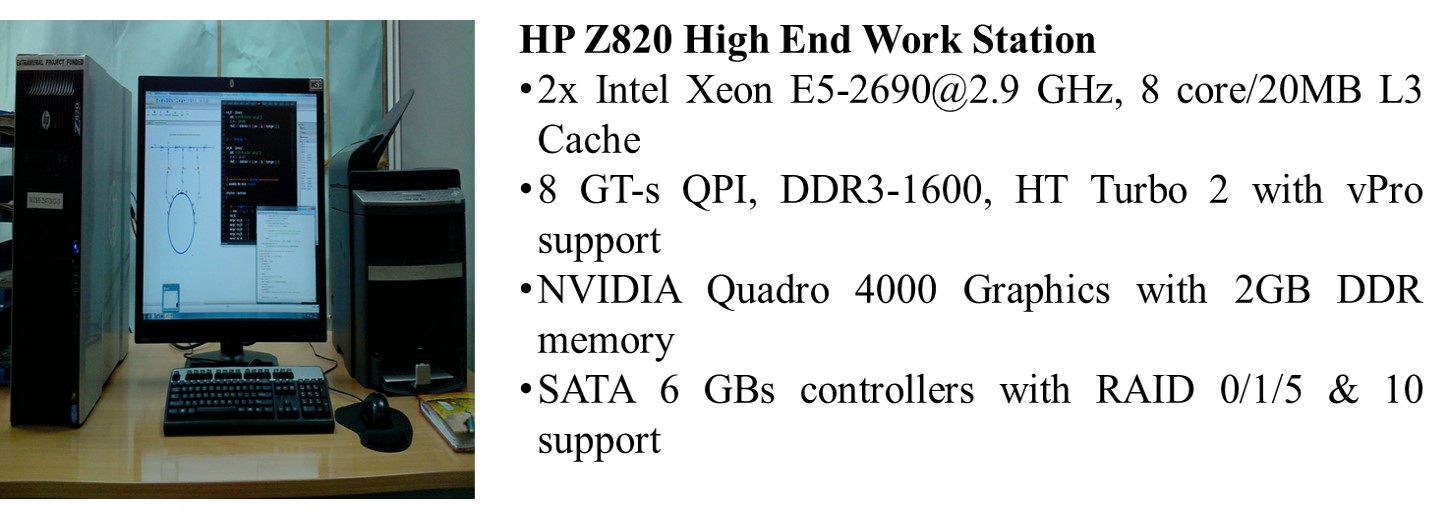
GPU Computing HP Proliant SL6500 SGI Altix XE 1300 Cluster |
2x Intel Xeon X5675 @3.06GHz/6 core/12MB L3 Cache 96 GB (8 GB x 12) PC3 – 10600 (DDR3 – 1333) Registered DIMM memory 2 x 1 TB hot Plug SATA Hard Disk @7200 rpm Integrated Graphics ATI RN50/ES1000 with 64 MB memory 2x NIVIDIA Tesla 2090 6 GB GPU computing module |
 |
|
iMAC: For running specialized software like Biojade APC UPS 10 KVA for supporting the HPCF |
|
 |
|
 |
|
|
|
|
Email: singhs@nccs.res.in
Phone: 020-25708295/8296