
Nibedita Lenka, Ph.D.
nibedita@nccs.res.in
Research Areas
Stem Cells and Regeneration
Education and Experience
1988-1992: Ph.D. (Genetics), Osmania University, Hyderabad, India.
1992-1993: Postdoctoral Research Associate, Temple University, Philadelphia, USA.
1993-1998: Postdoctoral Fellow, University of Pennsylvania, Philadelphia, USA.
1998-2001: Postdoctoral Research Scientist, University of Cologne, Germany.
2001-present: Scientist, National Centre for Cell Science, Pune, India.
Research
Embryonic stem (ES) cells being derived from the blastula stage embryo are capable of recapitulating the early embryogenic proceedings in a relatively precise manner, and hence serve as an elegant in vitro model to dissect out the developmental events occurring during embryogenesis. They possess the unique characteristics of indefinite self renewal and pluripotency. The major focus of our group has been the investigation of ES cells maintenance and differentiation into various cell types and the exploration of the molecular basis of cell commitment and differentiation into various lineages.
1. Factors influencing the maintenance of Embryonic Stem Cells in undifferentiated state
One of the most critical parameters in ES cells research is to maintain the cells in undifferentiated state. Failure to do so results in the loss of pluripotency. Accordingly, we have been investigating the influence of various signalling cascades (Wnt, Notch, PI3K) in parallel to the conventional LIF-Stat3, on murine ES cells?maintenance and the subsequent retention of pluripotency both in vitro and in vivo. Our investigation has revealed Wnt activation, though appears dispensable, it helps ES cells to remain undifferentiated even in the absence of LIF and the cells can be maintained in prolonged culture without losing their pluripotent characteristics. Conversely, while PI3K seems to be mandatory, Notch does not seem to have pronounced influence during ES cells maintenance.
2. Unraveling the complex temporo-spatial cell fate decision machinery from uncommitted ES cells
During the onset of gastrulation the commitment and specifications of various lineages take place. However, the question arises how does the cell decide which lineage to opt for? In fact, the cell intrinsic genetic make up and the extrinsic environment, both influence the cell fate decision machinery. Accordingly, we have aimed at understanding the cellular cross-talk and the key molecular determinants underlying the cell fate decision machinery specifically during early neuro- and cardiomyogenesis using ES cells as a model system. By following live reporter based cell trapping approach, we do demarcate the cells of interest ?ive?among the heterogeneous pool of differentiating ES cells. We have been successful in generating a number of stable ES cell clones that express the fluorescent reporter genes under the regulatory control of respective lineage specific promoters/enhancers. In fact, the said strategy has helped us in monitoring the developmental progression of cardiomyocytes, neural progenitors and dopaminergic neurons during differentiation from ES cells, and various projects are ongoing in understanding the underlying guiding cues dictating divergent cell fates decision.
(a) ES cells and Cardiomyogenesis
We have chosen the ES cell model to generate cardiomyocytes in vitro and have investigated the molecular basis of cardiac lineage specification and cardiomyocytes development. Following the promoter/enhancer mediated cell trap approach and using the fluorescent reporter system enabled us to demarcate effectively the cardiac population and to monitor their developmental progression "live" in culture. Accordingly, a number of cardiac specific stable clones have been generated that show spontaneous pulsating activity during differentiation with fluorescence remaining restricted to the beating areas only and hence proves the authenticity of these clones. The beating pattern in various areas remains either simultaneous or consecutive indicating the probable presence of different pace making centers. We have also been able to purify the cardiomyocytes employing genetic selection, which show expression of various cardiac transcription factors and the atrial, ventricular and conduction system specific genes. While investigating the guiding cues operational during early cardiomyogenesis, we have observed the activation of canonical Wnt signaling to inhibit cardiomyogenesis in a dose dependent manner indicating its threshold dependent negative influence on the cardiac differentiation process, Interestingly, the critical time window for its action resides at the early stages of differentiation. Conversely, BMP inhibition has led to cardiomyogenic abrogation, which however, is nullified with further inhibition of Wnt. Thus, our investigation has delineated an interesting synergy existing between Wnt and BMP and we have also demonstrated that, instead of all or none, it is the threshold of these two factors along with their temporal influence that guides the cardiomyogenic specification and development.
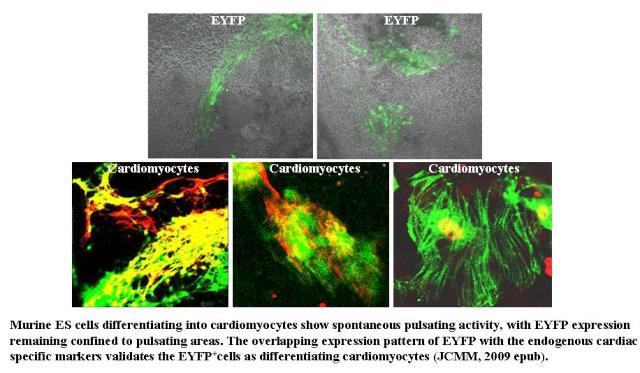 |
(b) ES cells and Neurogenersis
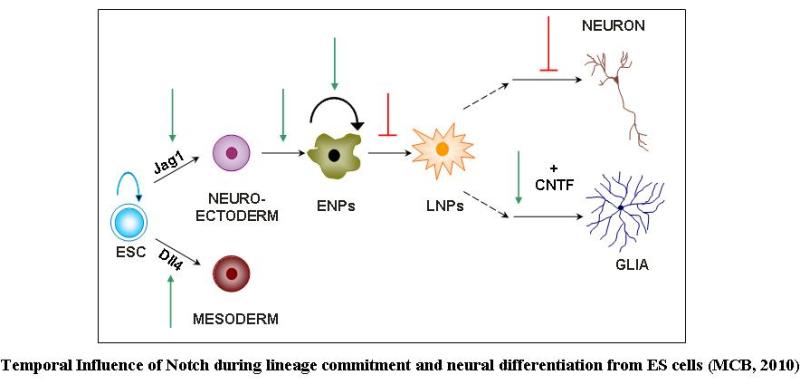
We have taken advantage of the neural specific expression of live reporters under the regulatory control of neural specific promoters/enhancers in ES cells in order to establish ES cell derived neural progeny and demarcate the cells of interest in the heterogeneous cell mass. Accordingly, we have chosen two different neural specific genes expressed at different times during embryonic development i.e. the early onset intermediate filament gene, the nestin, which is a marker for mitotically active neural stem cells/progenitors and the late expressed dopaminergic neuron specific gene, the tyrosine hydroxylase and have successfully generated a number of stable ES cell clones expressing the live reporter EGFP in neural progenitors and dopaminergic neurons respectively. Using nestin EGFP transgenic ES cell clones we have determined the optimum time window for neural progenitor generation that resides around one week of post-plating. During differentiation, the EGFP positive neural progenitors give rise to both MAP2 positive neuronal cells and GFAP positive astroglial cells. We have in fact, devised a monoculture strategy for neural differentiation in the absence of serum that yields ~80% differentiated neurons in culture. Further investigations have been carried out to asses the role of various factors during neural induction and subsequent differentiation. Our investigation has revealed a multifaceted role of Notch demonstrating the interdependency of ligand usage and lineage specification and Notch acting as a master switch displaying stage specific influence on neurogenesis. Similarly, using the TH transgenic ES cell clones we have been monitoring the developmental pathway leading to dopaminergic neuron generation. In parallel, we are investigating the therapeutic efficacy of ES cell derived neural progenitors and dopaminergic neurons by establishing hemi-parkinsonian rat models and injecting the stated in vitro generated cells into the brain using stereotaxy means.
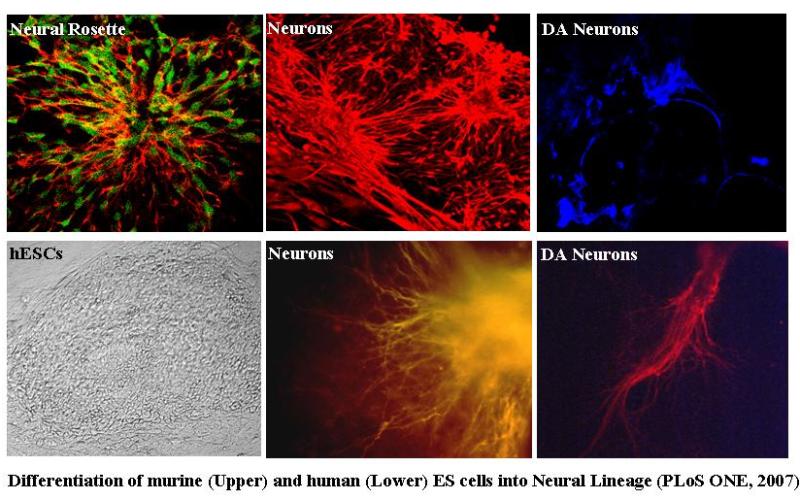 |
  |
3. Establishment of human ES cell line, characterization and differentiation
Attempts have been made in our laboratory, vide a DBT sponsored project, to establish human ES (hES) cell lines using the discarded human embryos from IVF clinics, as per the specified ethical guidelines. A couple of human embryos obtained were having three pronuclei, hence were considered unsuitable for implantation and were meant to be discarded. One of those successfully attained the blastocyst stage with a relatively indiscernible ICM. The ICM was isolated and upon culturing on mitotically inactive mouse embryonic fibroblasts (MEF), could give rise to hES cell like outgrowth. Subsequently this hES cell outgrowth could be propagated and maintained till P15 in culture. They showed the expression of various hES cells specific markers, however, exhibited a very high propensity towards spontaneous differentiation - a major bottleneck for retaining pluripotency. Concurrently, the cells were also induced to differentiate to determine their pluripotent status. We have succeeded in differentiating the hES cells into neural lineage. Concentric rings of skeletal muscle fibers were also seen in these cultures. The study overall indicated the established hES cells having the potential to differentiate into both ectodermal and mesododermal lineages.
4. Isolation of side population (SP) cells from umbilical cord blood and adult bone marrow. Expansion and phenotypic characterizations of MSCs and investigation of differentiation/ transdifferentiation potential, if any, into various lineages
We have carried out the investigations on mesenchymal stem cells (MSCs) from various sources like human bone marrow (hBM), umbilical cord (UC) and umbilical cord blood (UCB) to chalk out the strategies for their enrichment, and to explore their transdifferentiation potential, if any. Attempts have also been made to isolate and purify the side population (SP) cells, the characteristic phenotype seen in many adult stem cells/progenitors including the cancer stem cells. While both hBM and UC derived MSCs could be propagated in long term culture, UCB derived ones could not be passaged beyond P3-4. These cells were further immuno-phenotyped using various surface markers. The doubling time of these UC derived MSCs was comparatively less than the hBM ones and they could be successfully differentiated into osteocytes and adipocytes, displaying the inherent characteristics of MSCs. A number of reports indicate MSCs to be capable of transdifferentiation into various lineages. Hence, we intended to verify whether it does occur in vitro and whether we could transdifferentiate those into neural derivatives and cardiomyocytes by cultural manipulations. In our hand, we could not succeed in getting the pulsating cells from MSCs in culture upon 5-azacytidine treatment, the potent inducer of cardiomyocytes. We did observe nestin, Sox1 and Sox2 positive cells, the marker for neural progenitors and occasionally GFAP positive astroglial cells. However, a conclusive picture on transdifferentiation could not be drawn and further work is needed to authenticate this.
Patents
P. B. Parab, S. Ghaskadbi, Vidya Patwardhan, Nibedita Lenka, G. C. Mishra, Anil Chatterjee, P. S. Parameswaran. A new molecule for cardiac development promoting activity. Application No. 435/MVM/05/ Filing Date 6/04/2005, Indian Patent Office.
Publications
Publications
| K. Govarthanan, P. Vidyasekar, P. K. Gupta, N. Lenka, R. S. Verma. (2020) Glycogen Synthase Kinase 3β Inhibitor- CHIR 99021 Augments the Differentiation Potential of Mesenchymal Stem Cells., Cytotherapy, 22: 91-105. DOI: doi: 10.1016/j.jcyt.2019.12.007. |
| M. L. Desai, B. Deshmukh, N. Lenka, V. Haran, S. Jha, H. Basu, R. K. Singhal, P.K. Sharma, S. K. Kailasa, K. H. Kim. (2019) Influence of doping ion, capping agent and pH on the fluorescence properties of zinc sulfide quantum dots: Sensing of Cu2+ and Hg2+ ions and their biocompatibility with cancer and fungal cells., Spectrochim Acta A Mol. Biomol. Spectrosc., 210: 212-221. |
| V. Haran, N. Lenka. (2019) Deciphering the Epitranscriptomic Signatures in Cell Fate Determination and Development., Stem Cell Rev. Rep., 15: 474-496. DOI: doi.org/10.1007/s12015-019-09894-3. |
| A. Rajendran, U. Kapoor, N. Jothinarayanan, N. Lenka*, D. Pattanayak*. (2019) Effect of Silver-Containing Titania Layers for Bioactivity, Antibacterial Activity, and Osteogenic Differentiation of Human Mesenchymal Stem Cells on Ti Metal., ACS Appl. Bio Mater, 2: 3808-3819. DOI: DOI: 10.1021/acsabm.9b00420. |
| K. R. Aadil*, A. Nathani, C. S. Sharma*, N. Lenka*, P. Gupta*. (2019) Investigation of poly(vinyl) alcohol-gellan gum based nanofiber as scaffolds for tissue engineering applications., J. Drug Discov. Sci. Technol., 54: 101276. |
| J. Bhamore, B. Deshmukh, V. Haran, S. Jha, R. K. Singhal, N. Lenka, S. K. Kailasa, Z.V.P. Murthy. (2018) One-step eco-friendly approach for the fabrication of synergistically engineered fluorescent copper nanoclusters: Sensing of Hg2+ ion and cellular uptake and bioimaging properties., New J. Chem, 42: 1510-1520. |
| K. R. Aadil*, A. Nathani, C. S. Sharma, N. Lenka*, P. Gupta*. (2018) Fabrication of biocompatible alginate-poly(vinyl alcohol) nanofibers scaffolds for tissue engineering applications., Materials Technol., 33: 507-512. |
| C. Dey, G. Narayan, H. Krishna Kumar, MP. Borgohain, N. Lenka, RP. Thummer. (2017) Cell-Penetrating Peptides as a Tool to Deliver Biologically Active Recombinant Proteins to Generate Transgene-Free Induced Pluripotent Stem Cells., Stud. Stem Cells Res. Ther., 3: 06-15. |
| N. Lenka (2017) A Stem Cell Perspective on Cell fate Specification and Personalized Medicine., Insights on Global Challenges and Opportunities for the Century Ahead, Ed: V.D. Reddy, K.V. Rao, K.R, Osmania Univers: 203-214. |
| SL. D'souza B. Deshmukh JR. Bhamore KA. Rawat N. Lenka SK. Kailasa (2016) Synthesis of fluorescent nitrogen-doped carbon dots from dried shrimps for cell imaging and boldine drug delivery system, RSC Advances, 6: 12169-12179. PMID: 00 DOI: 10.1039/C5RA24621K |
| SL. D’souza, B. Deshmukh, KA. Rawat, N. Lenka, SK. Kailasa. (2016) Fluorescent carbon dots derived from vancomycin for flutamide drug delivery and cell imaging., New J. Chem., 40: 7075-7083. PMID: 0 DOI: 10.1039/C6NJ00358C |
| D. Rangasamy, N. Lenka, S. Ohms, J. E. Dahlstrom, A. Blackburn, P. Board. (2015) Activation of LINE-1 retrotransposon increases the risk of epithelial-mesenchymal transition and metastasis in epithelial cancer., Curr Mol Med., 15: 588-597. |
| N. Lenka, S. Krishnan, P. Board and D. Rangasamy (2014) Exploiting the power of LINE-1 retrotransposon mutagenesis for identification of genes involved in embryonic stem cell differentiation., Stem Cell Rev., 10: 408-416. |
| S. K. Ramasamy and N. Lenka. 2010. Notch Exhibits Ligand Preference and Maneuvers Stage Specific Steering of Neural Differentiation in ESCs. Mol. Cell. Biol 30:1946-1957. |
| N. Lenka and A. Anand. 2009. Advancements in Stem Cell Research - An Indian Perspective. Annals Neuro Sci. 16: 97-98 (Editorial). |
| M. K. Verma and N. Lenka. 2009. Temporal and Contextual Orchestration of Cardiac Fate by WNT-BMP Synergy and Threshold. (J. Cell. Mol. Med.: In Press). |
| S. Ghaskadbi, V. Patwardhan, M. Chakraborthy, S. Agrawal, M. K. Verma, A. Chatterjee, N. Lenka, P.B. Parab. 2008. Enhancement of Vertebrate Cardiogenesis by a Lectin from Perivitelline Fluid of Horseshoe Crab Embryo. Cell. Mol. Life Sci. 65:3312-3324. |
| N. Lenka, and S.K. Ramasamy. 2007. Neural induction from ES cells portrays default commitment but instructive maturation. PLoS ONE 2:e1349. |
| N. Lenka, G. C. Mishra. 2007. Stem Cells: The Present Status and the Future Prospects. J. Cell Tissue Res. (View Point: Invited). |
| N. Lenka. Derivation and Characterization of Neural Cells from Embryonic Stem Cells using Nestin Enhancer. 2006. Methods in Molecular Biology Vol. 330, Embryonic Stem Cell Protocols, Ed: K. Turksen, Humana Press, Second Edition, 2:33-54 (Invited Chapter). |
| N. Lenka. Investigation of early Cardiomyogenesis and Neurogenesis using ES cell model. 2005. Ranbaxy Science Foundation? 11th Annual Symposium Proceedings on ?tem Cell Research: From Bench to Bedside? Ed: O.P. Sood and R. S. Bakshi, pp 35-39. |
Other Publications
| B. K. Fleischmann, Y. Duan, Y. Fan, T. Schoeneberg, A. Ehlich, N. Lenka, S. Viatchenko-Karpinski, L. Pott, J. Hescheler, B. Fakler. 2004. Differential subunit composition of the G protein-activated inward-rectifier potassium channel during cardiac development. J. Clin. Invest. 114: 994-1001. |
| K. Govarthanan, P. Vidyasekar, P. K. Gupta, N. Lenka, R. S. Verma. (2020) Glycogen Synthase Kinase 3β Inhibitor- CHIR 99021 Augments the Differentiation Potential of Mesenchymal Stem Cells., Cytotherapy, 22: 91-105. PMID: |
| M. L. Desai, B. Deshmukh, N. Lenka, V. Haran, S. Jha, H. Basu, R. K. Singhal, P.K. Sharma, S. K. Kailasa, K. H. Kim. (2019) Influence of doping ion, capping agent and pH on the fluorescence properties of zinc sulfide quantum dots: Sensing of Cu2+ and Hg2+ ions and their biocompatibility with cancer and fungal cells., Spectrochim Acta A Mol. Biomol. Spectrosc., 210: 212-221. PMID: |
| V. Haran, N. Lenka. (2019) Deciphering the Epitranscriptomic Signatures in Cell Fate Determination and Development., Stem Cell Rev. Rep., 15: 474-496. PMID: |
| A. Rajendran, U. Kapoor, N. Jothinarayanan, N. Lenka*, D. Pattanayak*. (2019) Effect of Silver-Containing Titania Layers for Bioactivity, Antibacterial Activity, and Osteogenic Differentiation of Human Mesenchymal Stem Cells on Ti Metal., ACS Appl. Bio Mater, 2: 3808-3819. PMID: |
| K. R. Aadil*, A. Nathani, C. S. Sharma*, N. Lenka*, P. Gupta*. (2019) Investigation of poly(vinyl) alcohol-gellan gum based nanofiber as scaffolds for tissue engineering applications., J. Drug Discov. Sci. Technol., 54: 101276. PMID: |
| J. Bhamore, B. Deshmukh, V. Haran, S. Jha, R. K. Singhal, N. Lenka, S. K. Kailasa, Z.V.P. Murthy. (2018) One-step eco-friendly approach for the fabrication of synergistically engineered fluorescent copper nanoclusters: Sensing of Hg2+ ion and cellular uptake and bioimaging properties., New J. Chem, 42: 1510-1520. PMID: |
| K. R. Aadil*, A. Nathani, C. S. Sharma, N. Lenka*, P. Gupta*. (2018) Fabrication of biocompatible alginate-poly(vinyl alcohol) nanofibers scaffolds for tissue engineering applications., Materials Technol., 33: 507-512. PMID: |
| C. Dey, G. Narayan, H. Krishna Kumar, MP. Borgohain, N. Lenka, RP. Thummer. (2017) Cell-Penetrating Peptides as a Tool to Deliver Biologically Active Recombinant Proteins to Generate Transgene-Free Induced Pluripotent Stem Cells., Stud. Stem Cells Res. Ther., 3: 06-15. PMID: |
| N. Lenka (2017) A Stem Cell Perspective on Cell fate Specification and Personalized Medicine., Insights on Global Challenges and Opportunities for the Century Ahead, Ed: V.D. Reddy, K.V. Rao, K.R, Osmania Univers: 203-214. PMID: |
| SL. D'souza B. Deshmukh JR. Bhamore KA. Rawat N. Lenka SK. Kailasa (2016) Synthesis of fluorescent nitrogen-doped carbon dots from dried shrimps for cell imaging and boldine drug delivery system, RSC Advances, 6: 12169-12179. PMID: 00 |
| SL. D’souza, B. Deshmukh, KA. Rawat, N. Lenka, SK. Kailasa. (2016) Fluorescent carbon dots derived from vancomycin for flutamide drug delivery and cell imaging., New J. Chem., 40: 7075-7083. PMID: 0 |
| D. Rangasamy, N. Lenka, S. Ohms, J. E. Dahlstrom, A. Blackburn, P. Board. (2015) Activation of LINE-1 retrotransposon increases the risk of epithelial-mesenchymal transition and metastasis in epithelial cancer., Curr Mol Med., 15: 588-597. PMID: |
| N. Lenka, S. Krishnan, P. Board and D. Rangasamy (2014) Exploiting the power of LINE-1 retrotransposon mutagenesis for identification of genes involved in embryonic stem cell differentiation., Stem Cell Rev., 10: 408-416. PMID: |
| E. Boopathi, N. Lenka, S. K. Prabu, J. K. Fang, F. Wilkinson, M. Atchison, A. Giallongo, N. G. Avadhani. 2004. Regulation of murine cytochrome c oxidase Vb gene expression during myogenesis: YY1 and a CArG binding protein reciprocally regulate the transcription activity by physical interaction with BERF-1/ZBP-89 factor. J. Biol. Chem. 279:35242-35254. |
| S. Kazemi, D. Wenzel, E. Kolossov, N. Lenka, A. Raible, P. Sasse, J. Hescheler, K. Addicks, B. K. Fleischmann, W. Bloch. 2002. Differential Role of bFGF and VEGF for Vasculogenesis. Cell Physiol. Biochem. 12:55-62. |
| N. Lenka, Z. J. Lu, P. Sasse, J. Hescheler, B.K. Fleischmann. 2002. Quantitation and functional characterization of neural cells derived from ES cells using nestin enhancer mediated targeting in vitro. J. Cell Sci. 115:1471-1485. |
| C. Andressen, E. Stocker, F. J. Klinz, N. Lenka, J. Hescheler, B. Fleischmann, S. Arnhold, K. Addicks. 2001. Nestin-specific green fluorescent protein expression in embryonic stem cell-derived neural precursor cells used for transplantation. Stem Cells19:419-424. |
| J. Su, H. Yu, N. Lenka, J. Hescheler, S. Ullrich. 2001. Expression and regulation of depolarization-activated K+ channels in the insulin secreting cell line INS-1. Pfl\FCg. Arch. Eur. J. Physiol. 442:49-56. |
| S. M. Huber, B. Friedrich, K. Klingel, N. Lenka, J. Hescheler, F. Lang. 2001. Protein and mRNA expression of serum and glucocorticoid- dependent kinase 1 (SGK1) in metanephrogenesis. Dev. Dyn. 221:464-469. |
| C. Vijayasarathy, S. Damle, N. Lenka, and N. G. Avadhani. 1999. Tissue variant effect of Heme inhibitors on the mouse cytochrome c oxidase Gene expression and Catalytic Activity of the enzyme complex. Eur. J. Biochem. 266:191-200. |
| C. Vijayasarathy, I. Biunno, N. Lenka, Y. Ming, A. Basu, I. P. Hall, and N. G. Avadhani. 1998. Tissue and cardiac compartment specific variations in the composition and activity of the Cytochrome c Oxidase complex. Biochim. Biophys. Acta 1371:71-82. |
| N. Lenka, C. Vijayasarathy, J. Mullick, and N. G. Avadhani. 1998. Structural organization and transcription regulation of nuclear genes encoding the mammalian Cytochrome c Oxidase complex. Progress in Nuc. Acids Res. And Mol. Biol., (Ed: Kivie Moldave), Academic Press 61:309-344 (Invited Chapter). |
| A. Basu, N. Lenka, J. Mullick and N. G. Avadhani. 1997. Regulation of Murine Cytochrome Oxidase Vb Gene Expression in Different Tissues and During Myogenesis: Role of a YY-1 Factor Binding Negative Enhancer. J.Biol. Chem. 272:5899-5908. |
| N. Lenka, A. Basu, J. Mullick and N. G. Avadhani. 1996. The role of an E box binding Helix Loop Helix protein in the cardiac muscle-specific expression of the rat Cytochrome oxidase subunit VIII gene. J. Biol. Chem. 271:30281-30289. |
| N. Lenka and G.M.Reddy. 1994. Role of Media, Plant Growth Regulators in Callusing and Plant Regeneration from Anthers of Indica Rice. Proc. Indian Natl. Sci. Acad. 60:87-92. |
| N. Lenka, M. Aruna and G. M. Reddy. Genotypic differences and androclonal variation in anther culture of indica rice. Proc. Symp. Recent Advances in Rice Genetics and Tissue culture (Ed) G. M. Reddy, Hyderabad, India. |
| N. Lenka and G. M. Reddy. 1990. Seedling growth of indica rice under sea water stress. Proc.Symp.Rice Research: New Frontiers (Ed) E. A. Siddiq, Hyderabad, India, 414-415. |
Reviewer
Reviewed Manuscripts for Scientific Journals
Current Science
Stem Cells
Current Trends in Biotechnology and Pharmacy
Journal of Tissue Engineering and Regenerative Medicine
ICFAI Journal of BioTechnology
Aquaculture Research
Stem Cell Reviews and Reports
Indian Journal of Experimental Biology
Reviewed Research Projects for Funding Agencies
Department of Biotechnology (DBT), Department of Science and Technology (DST), Council of Scientific and Industrial Research (CSIR), KSCSTE, Tata Memorial Hospital (TMH), Indo-Australia, Indo-French, D. S. Kothari Fellowship Programme.
Evaluated Thesis submitted for Ph.D. in Genetics and Biochemistry.
Evaluated Thesis submitted for Ph.D. in Applied Biology.
Awards/Honours/Memberships
Member Editorial Advisory Board, Annals of Neurosciences (2009).
Visiting Scientist (Invited), JSPS-DST Exploratory Exchange under the Indo-Japan Co-operative Program between Japan Society for the Promotion of Science (JSPS), Japan and DST, India (2007).
Member (Expert), Faculty Selection Committee, All India Institute of Medical Sciences (AIIMS), New Delhi (2005).
Member, Scientific Programme Committee, 22nd Annual Conference of Indian Academy of Neuroscience, "Neuroscience from basic to clinic"(2004/5).
Samanta Chandra Sekhar Award for the year 2002 in Life Sciences, Orissa Science Academy (Conferred in 2005).
Served as a member for the sub-committee formulated by DBT for the site visit to CMC, Vellore in regard to monitoring the project submitted to DBT (2004).
Selected participant and Scholarship recipient for the NIH sponsored HEST (Human Embryonic Stem Cell Toolbox) Workshop at the University of Georgia (2004).
91st Indian Science Congress Award in recognition of Scientific Achievements (2004).
Member, Scientific Committee, Reliance Life Sciences, Mumbai (2003).
Scholarship recipient, Keystone Symposia 2001 - Pluripotent Stem Cells: Biology and Applications.
Travel Award
International Society for Stem Cell Research (ISSCR) travel award (2008).
International Society for Stem Cell Research (ISSCR) travel award (2005).
National Institute of Health (NIH), USA travel award (2003).
Rockefeller Foundation Award (1991).
Funding
The work has been supported by NCCS intramural funding and various project grants from Department of Biotechnology, Government of India.
Present and Past Students
- Ms. Ranju Nair, S.R.F. (Immigrated to Canada)
- Mr. Mahesh Kumar Verma, (Ph.D. degree awarded; currently working as a Senior Scientist at Connexious, Bangalore).
- Mr. Saravana Kumar Ramasamy (S.R.F)
- Ms. Amrita Rai, S.R.F. (Got attracted for a Ph.D. in Germany).
- Mr. Suresh Kumar, M.Sc. (Project Assistant; currently pursuing his Ph.D. in Germany).
- Mr. Anand Kumar Pande, M.Tech. (Project Trainee; currently pursuing his Ph.D. at IIT, Chennai).
- Ms. Atri Ta, M. Sc. (Project Trainee; currently pursuing her Ph.D. at NICED, Kolkata).
- Ms. Anamika Singh, M.Sc. (Project Trainee; currently pursuing her Ph.D. at IIT, Roorkie).
- Ms. Stuthi Pragna Sadanala, M.Sc. (Project Trainee; currently engaged in teaching).
- Mr. Mustansir Bhori, M.Tech. (Project Trainee; currently engaged in teaching Masters students).
- Mr. Swagat Kumar Das, M.Tech. (Project Trainee)
- Mr. V. Kartikeya, M.Sc. (Project Trainee; currently engaged in Teaching and Research at MIRM, Bangalore).
Back
Last updated On : 29 November 2024 04:47


