Radha Chauhan, Ph.D.
radha.chauhan@nccs.res.in
Research Areas
Macromolecular Structure and Function, Cell Organization and Function
Education and Experience
2012-present, Scientist-D/E, National Centre for Cell Sciences, Pune, India
2004-2012, Postdoctoral/Research Associate; The Rockefeller University, New York, USA
2003-2004, Senior Research Assistant; Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India.
1998-2003, Ph.D. Institute of Microbial Technology (Affiliated to JNU Delhi), Chandigarh, India
1995-1997, M.Sc (Biochemistry), Jiwaji University, Gwalior, MP India
Research Focus
The nuclear pore complexes (NPCs) embedded in nuclear membrane bilayer solely mediate transport of all kind of macromolecules between nucleus and cytoplasm, and regulate nearly most cellular processes such as gene expression, mitosis, cell differentiation etc. Additionally, alternations in NPC and its associated proteins have been linked to several human diseases, such as cancer, genetic disorders and viral diseases. The architecture of the NPC is evolutionarily conserved and is a highly modular structure.
The nuclear pore complexes (NPCs) embedded in nuclear membrane bilayer solely mediate transport of all kind of macromolecules between nucleus and cytoplasm, and regulate nearly most cellular processes such as gene expression, mitosis, cell differentiation etc. Additionally, alternations in NPC and its associated proteins have been linked to several human diseases, such as cancer, genetic disorders and viral diseases. The architecture of the NPC is evolutionarily conserved and is a highly modular structure. Each NPC is comprised of ~30 different proteins called nucleoporins (Nups) that are arranged in multiple copies to yield a size of 65 MDa (yeast) or 125 MDa (vertebrate). In order to understand the molecular mechanisms of NPC assembly formed by these ~30 Nups and its versatile functions, the high-resolution structures are highly desired but complexity and the size of the NPCs pose tremendous challenges. A rational strategy therefore would be to disintegrate the components of NPC based on their structural and functional specificity and employ integrative approaches to learn about the roles of Nups in NPC assembly and cellular physiology. Our laboratory routinely utilizes various structural biology tools such as X-ray crystallography, electron microscopy and spectroscopy methods and we work in collaboration with cell biologists to understand the versatile functions of NPCs, such as how Nups participate in nucleocytoplasmic transport, gene regulation and cell differentiation.

Ongoing Research Projects:
1. Biochemical reconstitution and structural studies of human nucleoporin sub-complexes of the NPC
We are pursuing biochemical reconstitution and atomic resolution 3D structure determination of human Nup93 subcomplex, also known as inner ring complex and Nup62 sub-complex (known as central transport channel, CTC). We deciphered precise interaction regions of mammalian Nup93 that can be biochemically assembled with ordered region of Nup62•Nup54•Nup58 complex and showed EM structure using negatively stained method. We also demonstrated how the dynamic nature of the CTC complex is conserved across the species yet maintaining the species specific features.
See Biochemistry 2017, Protein Science 2019, Protein Science 2020
We also successfully reconstituted mammalian Nup88•Nup62•Nup214 complex and revealed the scaffold/hub role of Nup62 coiled-coil mediated heterotrimer formation with other Nups and such heterotrimers can be recognized by Nup93 based on 3D shape (biorxiv 2022)
We use cryo-EM Single particle analysis based methodologies to determine atomic resolution structure of human Nups, such as Nup155 (Biorxiv 2021) that revealed a biochemical consequences of a genetic mutation (R391H) to cause atria fibrilliation.
2. Crosstalk of NPC with viral proteins
We are pursuing the physical interactions of human Nups with viral factors to elucidate the underlying role of the NPC in viral infections such as HIV-1. (Cells 2019)
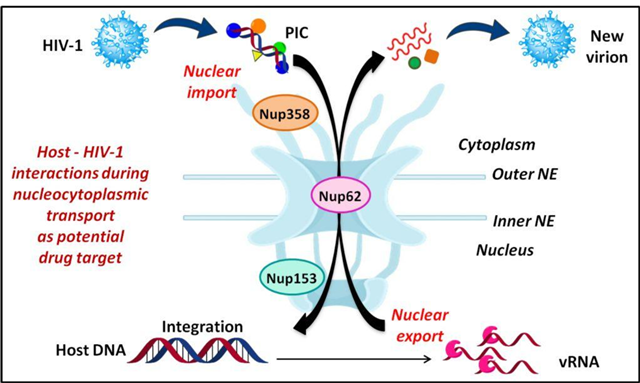
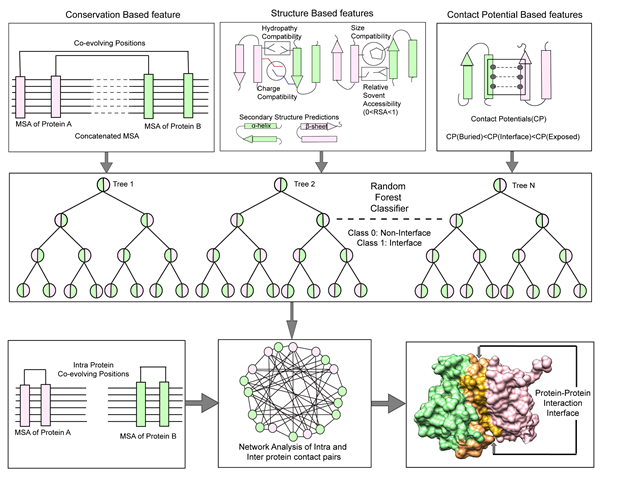
3. Developing innovative methods in protein-protein interactions
Using the combination of biochemical methods and machine learning computational tools we are actively pursuing development of new methods to decipher interacting network of multiprotein complexes. Our efforts in this area led us to come up with a tool (CoRNeA) that can predict interacting regions between two partners solely based on their primary sequences. Such method is extremely beneficial in biochemical reconstitution of the multimeric proteins (Biomolecules 2020).
OUR RESEARCH HIGHLIGHTS

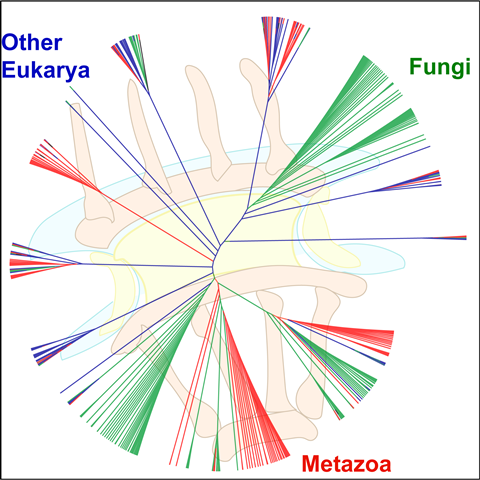
COVER PAGE OF BIOCHEMISTRY, 2017 Highlights of Protein Science, 2019 showing supertree of the NPC
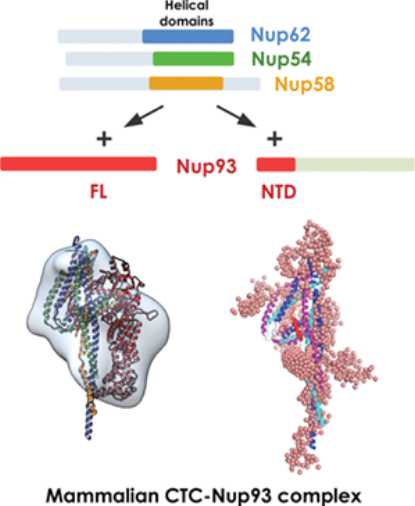
Highlights of Protein Science, 2020 issue showing architecture of mammalian CTC•Nup93 complex
Publications
2012 Onwards
| P. K. Madheshiya, E. Shukla, J. Singh, M. Y. Ansari, R. Chauhan. (2022). Insights into the role of Nup62 and Nup93 in assembling cytoplasmic ring and central transport channel of the nuclear pore complex (UNDER REVIEW) |
| S. Bawaria, P. J. Sonawane, R. Chauhan (2022). Structural Diversity of the Nuclear Pore Complexes (UNDER REVIEW) |
| S. Niranjan, J. Singh and R. Chauhan (2021). Cryo-EM structure of human Nup155 reveals the biochemical basis for atrial fibrillation linked genetic mutation R391H. biorxiv https://doi.org/10.1101/2021.10.05.463194. |
| S. Islam, P. Dutta, K. Chopra, O. Sahay, S. Rapole, R. Chauhan, M.K. Santra. 2021. Co-operative binding of SKP1, Cullin 1 and Cullin 7 to FBXW8 results in Cullin1-SKP1-FBXW8-Cullin7 functional complex formation that monitors cellular function of β-TrCP1. International Journal of Biological Macromolecules 190, 233-243. |
| B. George, A. Assaiya, R.J. Roy, A. Kembhavi, R. Chauhan, G. Paul, J. Kumar, N.S. Philip (2021). CASSPER is a semantic segmentation-based particle picking algorithm for single-particle cryo-electron microscopy. Communications Biology 4, 1-12. |
| S. Islam, P. Dutta, K. Chopra, S. Rapole, R. Chauhan, M.K. Santra. 2021. FBXW8 regulates G1 and S phases of cell cycle progression by restricting β‐TrCP1 function. The FEBS Journal ,10; doi:10.1111/febs.1582 |
| P. Sonawane*, P. Dewangan*, S. Neeranjan, P. Madheshiya, K. Chopra, S. Bawaria, M. Kumar, M. Banerjee and R. Chauhan 2020. Structural basis for Nup93 mediated anchoring of central transport channel of the nuclear pore complex. Protein Science 29(12) 2510-2527. Highlight of the October 2020 issue. |
| K. Chopra, B Budrak, K. Sharma, A. Kembhavi, S.C. Mande and R. Chauhan 2020. A pipeline to decrypt the protein-protein interaction from amino acid sequence information. Biomolecules 10(6). 938-951 |
| E. Shulka and R. Chauhan. 2019. Host-HIV-1 interactome: A quest for novel therapeutic interventions. Cells 8, 1155-1163. |
| K. Chopra, S. Bawaria and R. Chauhan (2019). Evolutionary divergence of the nuclear pore complex from fungi to metazoans. Protein Science 28(3); 571-586/. https://doi.org/10.1002/pro.3558. Highlight of the March 2019 issue. |
| P.S. Dewangan, P. J. Sonawane, A, R. Chouksey and R. chauhan (2017). The Nup62 coiled-coil motif provides plasticity for triple helix bundle formation, Biochemistry 56(22) 2803-2811. DOI: 10.1021/acs.biochem.6b01050. ‘COVER PAGE ARTICLE’. |
Before 2012:
| S.R. Solmaz*, R. Chauhan*, G. Blobel and I. Melćak (2011). The molecular architecture of the transport channel of the nuclear pore complex. Cell 147, 590-602. *Authors contributed equally to this work. |
| N. Kapoor, S.T. Menon, R. Chauhan, P. Sachdev and T.P. Sakmar (2009). Structural evidence for a sequential release mechanism for activation of hetero-trimeric G proteins.J Mol. Biol. 393, 882-897. |
| M. Akif, R. Chauhan and S.C. Mande (2004). Expression, purification, crystallization and preliminary X-ray crystallographic studies of Mycobacterium tuberculosis thioredoxin reductase. Acta Crystallogr D Biol Crystallogr. 60, 777-779. |
| Battu, S. Ghosh, A. K. Singh, S.C. Mande, V. Srinivas, R. Chauhan and N.Z. Ehtesham (2003). Human recombinant resistin protein displays a tendency to aggregate by forming intermolecular disulfide linkages. Biochemistry USA 42, 10554-10559. |
| C.W. Goulding, M. Apostol, D.H. Anderson, H.S. Gill, C.V. Smith, M.R. Kuo, J.K. Yang, G.S. Waldo, S.W. Suh, R. Chauhan, A. Kale, N. Bachawat, S.C. Mande, J.M. Johnston, J.S. Lott, E.N. Baker, V L. Arcus, D. Leys, K.J. McLean, A.W. Munro, J.Berendzen, V. Sharma, M.S. Park, D. Eisenberg, J. Sacchettini, T. Alber, B. Rupp, W. Jacobs, Jr. and T. C. Terwilliger (2002). The TB Structural genomics consortium: Providing a structural foundation for drug discovery. Curr. Drug Targets-Infect. Dis. 2, 121- 141. |
| R. Chauhan and S.C. Mande (2002). Site directed mutagenesis reveal a novel catalytic mechanism of Mycobacterium tuberculosis AhpC. Biochem. J. 267, 255-261. |
| R. Chauhan and S.C. Mande (2001). Biochemical characterization of Mycobacterium tuberculosis H37Rv points to the importance of ionic interactions in oligomerization and activity. Biochem. J. 354, 209-215. |
| B. Taneja, R. Chauhan and S.C. Mande (2000). Genomics and novel drug targets. Drugs and Pharmaceuticals Current R and D Highlights 23, 6-11. |
For latest publications :
Awards/Honours/Memberships
- DST/SERB Ramanujan fellowship 2012
- DST young scientist award 2003
- CSIR young scientist award 2002
- CSIR NET fellowship 1998-2003
Funding/Collaborations
Our laboratory research is funded by Department of Biotechnology and Department of Science and Technology India.
Radha Chauhan is Ramanujan Fellow
Lab Members
Virashree Jamdar, Technician C, virashree29@nccs.res.in
PhD Students
|
Pankaj Kumar Madheshiya |
2016-2022 |
|
Shrankhla Bawaria |
2016-2022 |
|
Manalee Thumpke |
2018- |
|
Jyotsna Singh |
2018- |
|
Aswathy LB |
2021- |
Postdoctoral Associate
| Priyanka Dutta, Inspire faculty 2018- |
| Ekta Shukla, DBT CoE-RA 2018- |
Alumni
| Graduate students | ||
| Pravin Dewangan | (2013-2019) | Postdoc fellow, Texas, USA |
| Kriti Chopra | (2014-2020) | Postdoc fellow BNL, USA |
| Bhawna Burdak | (2014-2021) | Scientist immunitoAI, Bangalore |
| Sangeeta Niranjan | (2015-2021) | Postdoc fellow, UCSF, USA |
| Pankaj Madheshiya | (2016-2022) | Thesis defense awaited |
| Postdocs | ||
| Nishant Varshney | 2013-2014 | Facility scientist, inSTEM, Bangalore India |
| Somnath Dutta, N-PDF | (2017-2019) | Scientist, Bharat Biotech India Ltd, Hyderabad |
| Parshuram Sonawane | (2014-2021) | Asst. Prof. Bharti Vidyapeeth, Pune |
| Ekta Shukla | (2018-2022) | Asst Manager, Gennova Biotech Pune |
B.S./M.Tech/M.Sc training students and project students trained in the lab:
| Name | Duration |
University |
| Moulshree Kulkarni | Jan’14-Jul’14 | Fakir Mohan university, Balasore, Orissa |
| Palak Agarwal | May’14-Jul’14 | University of Pune, Pune MH |
| I. Lawrence | May’14-Jul’14 |
Bharathidasan University, Triruchirapalli, TN |
| Jyoti Singh Gaharwar | Jan’15-May’15 | APS University of Rewa, MP |
| Kesha Dalal | May’15-Jul’15 | MS University of Baroda, Gujarat |
| Pushpanjali Rao | Jan’16-June’16 | Devi Ahilya University, Indore, MP |
| Sai N. Ramakrishnan | May-June 2016 | Sri S. Sai University, Bangalore, Karnataka |
| J Johnsi Rani | May-June 2017/td> | Sri Padmavathi womens college, Tirupathi AP |
|
Shrutika Sasane |
Jul’17-Dec’17 | Ahmednagar College, MH India |
| Somya Upadhyay | Feb ’18-Aug18 | Agra University, Agra, UP |
| Jyoti Sharma | Jan’19-June’19 | Himachal Pradesh University, HP |
| Simranjeet Kaur | Jan 20- June 20 | Punjab University, Chandigarh |
| Neda Mujaffar | Jan 21-June 21 | Jamia Millia Islamia, New Delhi |
|
Mrunmayee Ramteke |
Jan 21-June 21 | IBB, SPPU Pune |
| Siddhi Mukhedkar | Jan 22-June 22 | IBB, SPPU Pune |
| Suchitra Sharma | Feb 22-Jul 22 | DPSRU, New Delhi |
Opportunities:
Postdoctoral position: Highly motivated scientists interested in protein structure and function with strong background in cell biology, biochemistry and/or molecular biology or bioinformatics methods are always encouraged to apply. Inquire by email to Radha Chauhan
Ph.D positions: Prospective graduate students interested in our research (structural, computational and cellular biology) should apply through our regular Ph.D. program at NCCS and contact Radha Chauhan for possibilities of research projects.
2-12 months training: Students interested in MS/M.Sc project training projects, please email us at academics@nccs.res.in
Back
Last updated On : 24 August 2022 07:16


