
Dhiraj Dhotre, Ph.D.
Human microbiome, Role of microbiome in various metabolic diseases, Gut-brain axis, microbial genomics and metagenomics and computational biologydhiraj.dhotre@nccs.res.in
Research Areas
Microbial Ecology
Education and Experience
Education
Ph.D., 2014, National Centre for Cell Science, SP Pune University, Pune
M.Sc., 2005, Department of Bioinformatics, SP Pune University, Pune
B.Sc., 2003, B. N. Bandodkar College of Science, Mumbai University, Thane
Experience
Scientist D, 2021-present, National Centre for Cell Science, Pune.
Senior Research Scientist, 2019-2021, Reliance Life Sciences Pvt. Ltd., Navi Mumbai
Project Scientist C, 2017-2019, National Centre for Microbial Resource, National Centre for Cell Science, Pune
Project Scientist B, 2009-2017, Microbial Culture Collection, National Centre for Cell Science, Pune
Research
Soon after the birth, we acquire our microbiota that stabilizes over the next three years. Consequently, the adult human body becomes the home for complex microbial communities from all four domains of life (Bacteria, Archaea, Eukarya and Viruses) which are commonly referred to as ‘human microbiota’ while the genetic repertoire of these microbes together is referred to as ‘human microbiome’. As a result of this microbial succession and acquisition, trillions of microbes encompassing several different phyla, live in and on the human body throughout of our life. Of the various bodily sites, greatest numbers of microbes that are found in our distal gut that is known to affect literally every aspect of our lives.
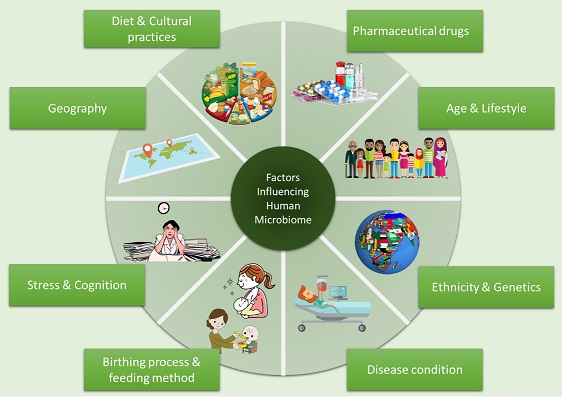
Concomitant with the development of next generation DNA sequencing technologies, we have succeeded in accomplishing much of the data regarding the structure of gut microbiota. Consequently, studies in past few years have highlighted discernible patterns of gut microbiome in plethora of diseases including neurological, immunological and metabolic to various life style associated disease such as cardiovascular, obesity, type 2 diabetes, cancer etc. Such studies have provided wealth of information related to peculiar gut microbial features and also have highlighted the need for population-specific studies to solve the issues related to the health of mankind. Hence, in order to tackle some of the country specific issues gut microbiota has to be given a due considerations. Our lab focuses on understanding the role of various confounding factors (such as diet, geography, age, and various metabolic and neurological disorders) in shaping the microbiome, especially from the Indian perspective.
Human Microbiome Initiative in select endogamous populations
Mounting evidence has been gathered suggesting that the commensal microbiota is important for the general wellbeing and its influence upon host physiology is increasingly recognized both locally and systemically. Perhaps the most important realization about the microbiota is that the human associated microbes are crucial component of host-microbe ecosystems that provides various services the host. Interestingly, the studies on indigenous microbial communities has also changed clinicians' perceptions about microbes from the causative agents to diseases to largely beneficial microbial communities that occupies several human body parts. Thus, it seems that there are no areas of human health or for that matters human diseases that have not been linked with the gut microbiota in one or the other ways. Initiation of many mega projects worldwide and the fact that human microbiota varies geographically, the commencement of nationwide Human Microbiome Initiative (HMI) was a need of time.
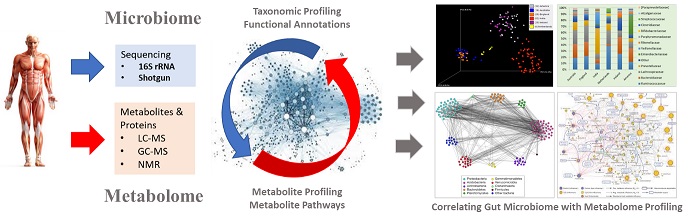
(HMI) is one of the ambitious and flagship programs of NCCS and DBT, Govt. of India. The human microbiome study will be carried out on 3400 individuals at two time points from various endogamous groups and tribal communities spread across the country. Targeted amplicon and metagenomic sequencing approaches are being used to study the microbiome from the gut. Bacterial, Fungal, Archaeal and Micro eukaryotic diversity will be captured in targeted amplicon approach using four independent DNA sequencing libraries. A correlation based on genotyping data generated using whole-genome sequencing will be drawn subset of individuals. We have also proposed extensive culturomics on a fraction of these samples, and the potential novel or important cultures will be genome sequenced. The urine metabolome will also be studied to understand its impact on human health and correlate it with the microbiome data generated. IHMI is a multi-institutional project led and coordinated by our group at NCCS.
Gut-Brain Axis
During the last decade or so, clinical as well as animal studies linked gut microbiota with many psychiatric disorders. Gut microbial dynamics is an enormously strong, intrinsic, biologically active non-genetic factor. It may be suggested that the microbiota may undertake epigenetic events through its impact on metabolism. In the current study, we are aiming to investigate the possibility of gut microbial changes affecting the brain epigenetic events which could be leading to regulation of downstream target genes implicated in neuronal communication and synaptic plasticity using rodent animal model. With the importance of epigenetics in shaping the experience-induced brain functions, it is paramount to establish the link, in gut microbiota and brain epigenetics. Our goal is to identify the epigenetic signatures of neuronal plasticity drawn in by gut microbiota with potential probiotic characteristics in the neurocircuitry of emotion and cognition, i.e., amygdala and hippocampus. Whole genome-wide epigenetic changes and whole metagenomic sequencing will be employed to establish the correlations in the gut microbiome and epigenetic changes in the brain of behaving animals. In addition, the observations drawn through these studies will be further employed to draw the mechanistic molecular model operative in behavioural implications of probiotics (Fig. 1).
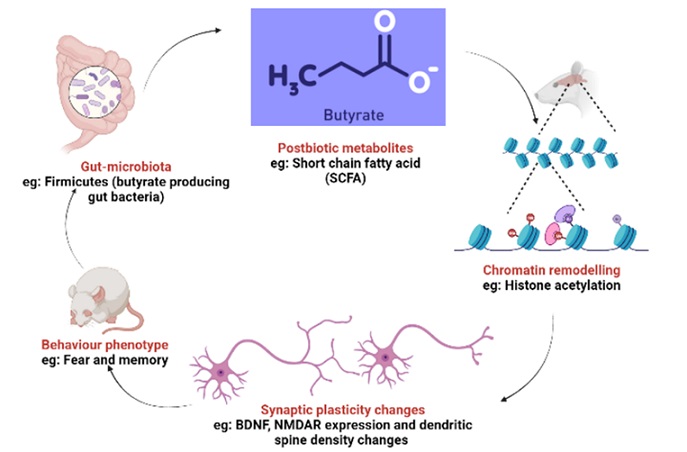
Schematic representation showing the hypothetical model of gut microbiota-brain-behaviour axis. Specific consortia of probiotic may enhance specific metabolites such as butyrates, propionates etc. (short chain fatty acids) in brain, which in turn can impact the chromatin remodelling of the genes involved in synaptic plasticity. These events may lead to behavioural changes and further create a cyclical axis of gut microbiota and behaviour.
Mother-Infant Microbiome and implications on infant health
The development of microbiome in infant occurs through mother-infant axis during or after the birth. First few days of life represent a crucial window of opportunity for shaping the development of the gastrointestinal tract and immune system, as well as the adult microbiome. But any dysbiosis during the development of microbiome in new-born can cause IBD, obesity, diabetes, inflammatory bowel disease and other long-term lifestyle disorders. The initial exposure of microbes in infant is said to begin during pregnancy through intra-uterine passage (fetal membranes, umbical cords, amniotic fluids) followed by the mode of delivery and by the type of feeding (breast feeding/ formulae fed) given to the infant. Breast milk is one of the continuous sources of mutually beneficial bacteria that are hypothesized to seed the infant’s gut via breastfeeding. It is believed that bacteria from mother’s gut translocate to breast milk via an entero-mammary pathway and thereby populate the infant’s oral and gut subsequently. Whereas, another hypothesis suggests the retrograde flux where a backward movement of milk occurs due to infant suckling and transfers the bacteria from infant’s mouth into the mother’s mammary gland. Mother’s breast milk serves as an important component in shaping the health of infant short term and long term as it harbours ideal balance of nutrients and the abundance of microbes which further acts as perfect food and an optimum medium for growth and development of immunity. Among all bioactive components present in milk, HMOs (Human Milk Oligosaccharides) presents chemical, microbial and medical importance. HMOs form a category of nondigestible, unconjugated, multifunctional and structurally glyans which resist gastrointestinal hydrolysis by gastric acidity and are not absorbable in significant amounts. Thus, allows HMOs to reach infant gut and produce a variety of beneficial effects. HMO’s acts as substrate for various useful bacteria like Bifidobacterium sp. where they are fermented in intestine. Apart from the nutritional role of HMOs in the intestinal microbiota of infants, there is increasing evidence that their interaction with pathogens is important in terms of reducing infection, for example HMOs exhibit bacteriostatic properties against group B Streptococcus (GBS). HMOs play a valuable role in development and maturation of the immune response by activating the expression of multiple cytokines and chemokines, adhesion molecules and receptors, thus maturing the naive immunity in infants. Any deviant in early microbial colonization in new-borns may lead to autoimmune diseases such as diabetes, inflammatory bowel disease, atopy, and other severe health conditions. It is known, that mother microbiome serves as reservoir in transmitting the microbes to infants. However, the exact source and route of infant pioneering microbes are poorly understood. Hence, to address this we propose a study of how different body sites of mother help in shaping the early microbiome of infant. The proposed longitudinal study will compare the effect of different maternal microbial communities (gut, breast milk and areolar skin) on the development of infant’s gut and oral
Moreover, to address this we have proposed an animal study of how different body sites of mother Balb/c mice help in shaping the early microbiome of infant mice. The proposed longitudinal study will compare the effect of different maternal microbial communities (gut, breast milk and areolar skin) on the development of infant’s gut and oral microbiome through high throughput species level targeted metagenomics (16S rRNA gene sequencing). For this purpose, fluorescently tagged specific bacterial stains will be transformed with a plasmid and subsequently, the transformed strains will be orally administered to pregnant mice. This mice model will allow the visualization, isolation, and detection of the transformed bacteria in different body locations, including mammary tissue and milk and hence reinforcing the hypothesis that physiological translocation of maternal bacteria during pregnancy and lactation may contribute to the composition of the microbiota of infant mice.
Gastrointestinal Disorders and Microbiome
CeD (Celiac disease) and NCGS (Non-celiac gluten sensitivity) disorders are still in infancy and consequently, straightforward diagnosis and treatment of both the diseases are yet to be delineated. Recently, there has been considerable attention on role of virome, mycobiome, archaea and micro-eukaryotes along with bacteriome various gastrointestinal disorders including GrDs (Gluten related disorders) pathogenesis. A recent study on Danish gut viromes generated Danish Enteric Virome Catalogue (DEVoC) through which numerous previously uncharacterized viral genomes and genes (12,986 nonredundant viral scaffolds encoding 190,029 viral genes) were added to the ever-increasing worldwide pool of human gut viromes. They have also proposed a potential role of virome in pathogenesis.
The proposed study aims to apply a holistic approach to characterize gut microbiome by employing taxonomic profiling (bacterial, fungal, archaeal and micro-eukaryotic diversity) and functional profiling (metagenome shotgun sequencing, virome sequencing, and meta-metabolomics) in individuals/patients suffering from different GrDs in correlation to self-declared healthy individuals. Such an extensive study, profiling all kinds of microbes, to characterize the gut microbiome of CeD and NCGS patients has not been carried out to the best of our knowledge. This approach of holistic gut microbiome characterization in conjunction with meta-metabolomic studies along with a focus on isolating gliadin degrading microbes will help us enhance our understanding of GrDs. This work will be an extension of two major initiative taken by Department of Biotechnology, Govt. Of India and will help us leverage huge amount of information and samples collected by Celiac disease consortium and biorepository project and Indian human microbiome project.
Colorectal Cancer and Microbiome
Colorectal cancer (CRC) is one of the most common cancers and ranks third in terms of incidence and second in mortality worldwide. The incidence of CRC is increasing in adults younger than 50 years of age. CRC etiology is highly complex and involves both genetic and environmental factors. Evidence from family studies indicates that only a small fraction of CRCs are genetically predispose. In addition, most CRCs are non-inherited. Environmental factors, such as western dietary habits, smoking, weight gain and obesity, diabetes, and heavy alcohol consumption, play major roles in causing sporadic CRC. Among environmental risk factors, the intestinal microbiota is an important contributor.
Growing evidence suggests that a state of pathological microbial imbalance or dysbiosis is prevalent in the gut of patients with colorectal cancer. Role of CRC-associated bacteria have been reported. Bacteria may affect CRC directly or indirectly, by secreting metabolites, by invading tissues, and by modulating the host immune response. Several bacterial taxa like Fusobacterium, Peptostreptococcus, Porphyromonas, Prevotella, Parvimonas, Bacteroides, and Gemella are among the most prominent CRC-associated bacteria which have been identified to interact with human cancer cells in vitro and trigger disease pathways in animal models.
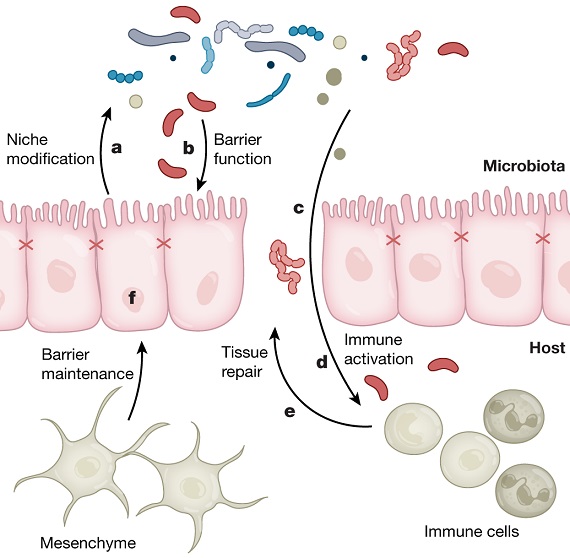
Currently, single microorganisms, consortia of microbes, and microbial metabolites have been identified that are being developed to improve cancer treatment efficacy. Interventions targeting the microbiota and cancer frequently mirror the spectrum of preventive and therapeutic strategies for infectious diseases, including vaccines, antibiotics, fecal microbiota transplants (FMTs), and phage-based therapeutics. A recent proof- of-principle study demonstrated that the antibiotic metronidazole could slow the growth of F. nucleatum positive human tumor samples in patient-derived xenograft mouse models. However, it is unclear whether antibiotics could be used in CRC prevention, given the years to decades over which cancerous lesions in the colon develop and the consequent potential of developing antibiotic resistance. Highly selective antibiotics or antivirulence approaches might represent a more viable strategy that is less disruptive for human microbial ecology.
FMT, the transfer of fecal material from healthy individuals to patients, is increasingly being used to combat colitis caused by antibiotic-resistant Clostridium difficile infection. More recently, FMT has been considered for the treatment of a number of diseases, including obesity and inflammatory bowel disease, with several clinical trials under way. FMT has also been piloted in a small number of cancer patients who developed severe colitis associated with the use of immunotherapy. Beyond FMT, carefully curated cocktails of microbes are being tested for C. difficile colitis and for CRC. The goal of such a therapeutic is to use microbial consortia to “push out” or exclude a disease-associated microorganism from a patient’s gut or tumor. Additionally, exciting opportunities also exist for using microbiota profiling information not only in CRC prevention, diagnostics, and therapeutics but also for treatments targeting GI microorganisms to decrease toxicities of CRC therapeutics.
Webservers and Computational tools
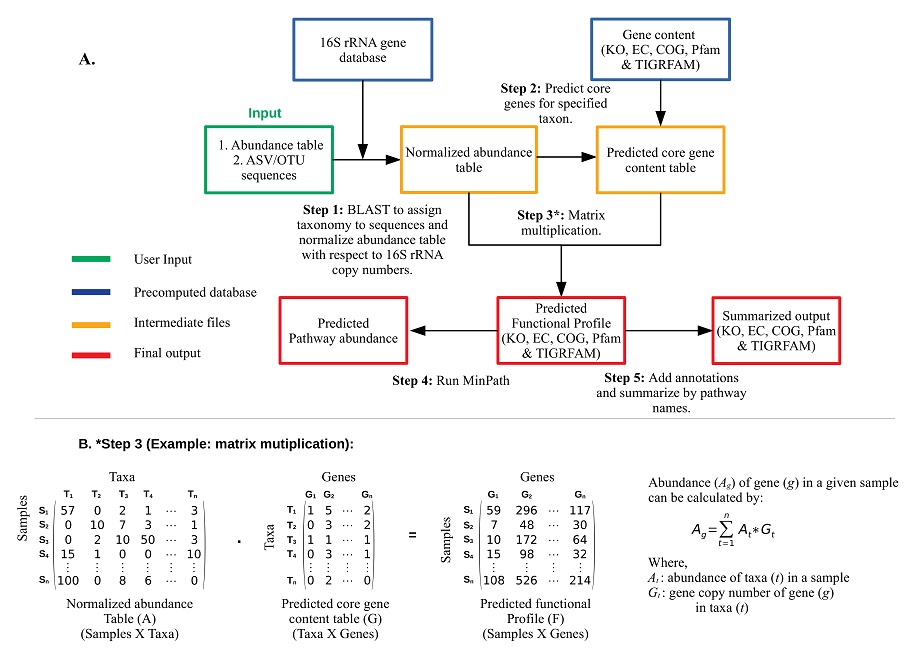
Our lab is actively involved in developing computational tools for answering most intriguing and complex questions in microbiome studies. ‘MicFunPred’ uses a set of core genes predicted at the genus level to derive imputed metagenomes with minimal false positive predictions. MicFunPred predicts a set of core genes using ~32,000 reference IMG permanent/finished draft genomes. On simulated datasets, MicFunPred showed the lowest False Positive Rate (FPR) with mean Spearman’s correlation of 0.89 (SD=0.03) while on 7 different real datasets the mean correlation was 0.75 (SD=0.08). MicFunPred was found to be faster with low computational requirements and performed better or comparable when compared with other tools. Webserver: http://micfunpred.microdm.net.in/ GitHub repo: https://github.com/microDM/MicFunPred
MicFunPred predicts functional profiles from 16S rRNA gene amplicon sequence data by using a more conserved approach involving core genes of each genus, thereby minimizing the false-positive results. An abundance table and 16S rRNA gene sequences from OTU/ASV based approach can be used directly. First, OTU/ASV sequences are searched against a custom 16S rRNA gene database using BLAST. The taxonomy is assigned to each OTU/ASV at the genus level (if percent identity >= user cutoff) and the abundance table is then consolidated at assigned taxonomy level. The consolidated abundance table then normalized with mean 16S rRNA gene copy numbers for each taxon. The gene content (only core genes) for each genus listed in the abundance table is predicted. Core genes are defined as a set of all genes present in x% of genomes from each genus (the value of ‘x’ can be adjusted by the ‘Gene coverage’ cut-off). Using normalized abundance and predicted gene content table, metagenomes in terms of KO, EC, TIGRFAM, COG, CAZymes & Pfam are calculated by performing a dot product of two matrices. For pathway prediction, naive approaches can give inflated estimates of biological pathways. Hence, we have used MinPath, which gives estimates of a minimal set of pathways using a more conservative parsimony approach.
ProbioPred
Probiotics, live microorganisms with beneficial health effects have become increasingly popular in the last two decades. Traditional approaches for screening and identifying potential probiotic strains are costly and time-consuming. Genome-based predictions using machine learning approaches have been implemented on a variety of phenotypic predictions. Here, we developed a user-friendly web-accessible application “ProBioPred” that can predict the probiotic potential of a candidate strain based on its genome. The method uses trained Support Vector Machine (SVM) models based on manually curated features like genes encoding probiotic properties, presence/absence of virulence factors and antibiotic resistance. Currently, ProBioPred supports the prediction of probiotic strains belonging to genera, Bacillus, Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, and Streptococcus due to their wide use as probiotics. The accuracy of trained models on test data for different genera ranged from 98% to 99.67% whereas the sensitivity and specificity range from 0.79 to 0.99 and 0.87 to 0.99 respectively. Models for each genus were validated on an independent dataset consisting of newly sequenced genomes with minimum completeness of 60%. ProBioPred is available with a graphical user interface that facilitates easy browsing and downloading of data. With higher accessibility and decreasing costs of next generation sequencing, ProBioPred can be a first step for prediction of potential probiotic bacteria before characterization of strains.
Availability: The ProBioPred web service is available at http://probiopred.in/ and as standalone at https://github.com/microDM/ProBioPred
Publications
Selected Publications All Publications
Selected Publications
-
Spehlmann ME, Rangrez AY, Dhotre DP, et al. Heart Failure Severity Closely Correlates with Intestinal Dysbiosis and Subsequent Metabolomic Alterations. Biomedicines. 2022;10(4):809.
-
Mongad DS, Chavan NS, Narwade NP, Dixit K, Shouche YS, Dhotre DP. MicFunPred: A conserved approach to predict functional profiles from 16S rRNA gene sequence data. Genomics. 2021;113(6):3635-3643.
-
Chaudhari DS, Dhotre DP, Agarwal DM, et al. Gut, oral and skin microbiome of Indian patrilineal families reveal perceptible association with age. Scientific Reports. 2020;10(1):1-13.
-
Gaike AH, Paul D, Bhute S, et al. The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. Msystems. 2020;5(2):e00578-19.
-
Alai S, Ghattargi VC, Gautam M, et al. Comparative genomics of whole-cell pertussis vaccine strains from India. BMC Genomics. 2020;21(1):1-15.
All Publications
-
Ghattargi VC, Shouche YS, Dhakephalkar PK, et al. Probiotics: A Mainstream Therapy for the Disease Suppression. 2022.
-
Sagar I, Nimonkar Y, Dhotre D, et al. A Microcosm Model for the Study of Microbial Community Shift and Carbon Emission from Landfills. Indian J Microbiol. 2022:1-9.
-
Spehlmann ME, Rangrez AY, Dhotre DP, et al. Heart Failure Severity Closely Correlates with Intestinal Dysbiosis and Subsequent Metabolomic Alterations. Biomedicines. 2022;10(4):809.
-
Rahi P, Chaudhari D, Kiran S, et al. Prokaryotic communities adapted to microhabitats on the Indian lotus (Nelumbo nucifera) growing in the high-altitude urban Dal Lake. 2022.
-
Bano S, More S, Mongad D, et al. Prolonged exposure to insulin causes epigenetic alteration leading to insulin resistance. bioRxiv. 2022.
-
Agarwal DM, Dhotre DP, Kumbhare S V, et al. Disruptions in oral and nasal microbiota in biomass and tobacco smoke associated chronic obstructive pulmonary disease. Arch Microbiol. 2021;203(5):2087-2099.
-
Assad R, Reshi ZA, Mir SH, Rashid I, Shouche Y, Dhotre D. Bioprospecting appraisal of Himalayan pindrow fir for pharmacological applications. In: Phytomedicine. Academic Press; 2021:461-482.
-
Kaushik R, Pandit MK, Meyerson LA, et al. Contrasting Composition, Diversity and Predictive Metabolic Potential of the Rhizobacterial Microbiomes Associated with Native and Invasive Prosopis Congeners. Curr Microbiol. 2021;78(5):2051-2060.
-
Shaligram S, Narwade NP, Kumbhare S V, et al. Integrated Genomic and Functional Characterization of the Anti-diabetic Potential of Arthrobacter sp. SW1. Curr Microbiol. 2021;78(7):2577-2588.
-
Dixit K, Chaudhari D, Dhotre D, Shouche Y, Saroj S. Restoration of dysbiotic human gut microbiome for homeostasis. Life Sci. 2021;278:119622.
-
Dixit K, Davray D, Chaudhari D, et al. Benchmarking of 16S rRNA gene databases using known strain sequences. Bioinformation. 2021;17(3):377.
-
Mane S, Dixit KK, Lathwal N, et al. Rectal administration of buttermilk processed with medicinal plants alters gut microbiome in obese individuals. J Diabetes Metab Disord. 2021;20(2):1415-1427.
-
Mongad DS, Chavan NS, Narwade NP, Dixit K, Shouche YS, Dhotre DP. MicFunPred: A conserved approach to predict functional profiles from 16S rRNA gene sequence data. Genomics. 2021;113(6):3635-3643.
-
Joshi A, Thite S, Dhotre D, et al. Nitrincola tapanii sp. nov., a novel alkaliphilic bacterium from An Indian Soda Lake. Int J Syst Evol Microbiol. 2020;70(2):1106-1111.
-
Kirdat K, Tiwarekar B, Thorat V, et al. Draft genome sequences of two phytoplasma strains associated with sugarcane grassy shoot (SCGS) and bermuda grass white leaf (BGWL) diseases. Mol Plant-Microbe Interact. 2020;33(5):715-717.
-
Jena J, Narwade N, Das T, Dhotre D, Sarkar U, Souche Y. Treatment of industrial effluents and assessment of their impact on the structure and function of microbial diversity in a unique Anoxic-Aerobic sequential batch reactor (AnASBR). J Environ Manage. 2020;261:110241.
-
Chaudhari DS, Dhotre DP, Agarwal DM, et al. Gut, oral and skin microbiome of Indian patrilineal families reveal perceptible association with age. Sci Rep. 2020;10(1):1-13.
-
Gaike AH, Paul D, Bhute S, et al. The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. Msystems. 2020;5(2):e00578-19.
-
Alai S, Ghattargi VC, Gautam M, et al. Comparative genomics of whole-cell pertussis vaccine strains from India. BMC Genomics. 2020;21(1):1-15.
-
Assad R, Reshi ZA, Rashid I, Shouche Y, Dhotre D. Role of ectomycorrhizal biotechnology in pesticide remediation. In: Bioremediation and Biotechnology, Vol 3. Springer, Cham; 2020:315-330.
-
Chaudhari DS, Dhotre DP, Jani K, et al. Bacterial Communities Associated with the Biofilms Formed in High-Altitude Brackish Water Pangong Tso Located in the Himalayan Plateau. Curr Microbiol. 2020. doi:10.1007/s00284-020-02244-4
-
Bodkhe R, Shetty SA, Dhotre DP, et al. Comparison of Small Gut and Whole Gut Microbiota of First-Degree Relatives with Adult Celiac Disease Patients and Controls. Front Microbiol. 2019;10(February):227272. doi:10.3389/fmicb.2019.00164
-
Kulkarni AS, Kumbhare S V, Dhotre DP, Shouche YS. Mining the Core Gut Microbiome from a Sample Indian Population. Indian J Microbiol. 2019;59(1):90-95.
-
Alai S, Ghattargi VC, Gautam M, et al. Genome sequence of bordetella pertussis vaccine strain BP 165. Microbiol Resour Announc. 2019;8(16). doi:10.1128/MRA.00150-19
-
Auti AM, Narwade NP, Deshpande NM, Dhotre DP. Microbiome and imputed metagenome study of crude and refined petroleum-oil-contaminated soils: Potential for hydrocarbon degradation and plant-growth promotion. J Biosci. 2019;44(5). doi:10.1007/s12038-019-9936-9
-
Chaudhari D, Dhotre D, Agarwal D, et al. Understanding the association between the human gut, oral and skin microbiome and the Ayurvedic concept of prakriti. J Biosci. 2019;44(5). doi:10.1007/s12038-019-9939-6
-
Matharu D, Dhotre D, Balasubramanian N, Pawar N, Sagarkar S, Sakharkar A. Repeated mild traumatic brain injury affects microbial diversity in rat jejunum. J Biosci. 2019;44(5). doi:10.1007/s12038-019-9940-0
-
Mane S, Dixit K, Lathwal N, et al. Per rectum administration of buttermilk processed with medicinal plants alters gut microbiota in obese individuals. In: JOURNAL OF GASTROENTEROLOGY AND HEPATOLOGY. Vol 34. WILEY 111 RIVER ST, HOBOKEN 07030-5774, NJ USA; 2019:272.
-
DHOTRE DP, KUMBHARE S V, SINKAR VP, SHOUCHE YS. Human Gut Microbiome Research in India: A Retrospect and Future Opportunities. In: Proc Indian Natn Sci Acad. Vol 85. ; 2019:1051-1066.
-
Bagade A V, Paul D, Giri A, Dhotre D, Pawar S, Kodam K. Arsenic oxidation by hypertolerant Bacillus sp. L-148 in artificial groundwater microcosm. In: Environmental Arsenic in a Changing World. CRC Press; 2019:587-588.
-
Jani K, Dhotre D, Bandal J, et al. World’s largest mass bathing event influences the bacterial communities of Godavari, a Holy River of India. Microb Ecol. 2018;76(3):706-718.
-
Ghattargi VC, Gaikwad MA, Meti BS, et al. Comparative Genome Analysis Reveals Important Genetic Factors Associated with Probiotic Property in Enterococcus faecium strains. bioRxiv. 2018:295881.
-
Ghattargi VC, Gaikwad MA, Meti BS, et al. Comparative genome analysis reveals key genetic factors associated with probiotic property in Enterococcus faecium strains. BMC Genomics. 2018;19(1):652. doi:10.1186/s12864-018-5043-9
-
Sharma A, Paul D, Dhotre D, Jani K, Pandey A, Shouche YS. Deep sequencing analysis of bacterial community structure of Soldhar hot spring, India. Microbiology. 2017;86(1):136-142. doi:10.1134/S0026261717010118
-
Chaudhari AUAU, Paul D, Dhotre D, Kodam KMKM. Effective biotransformation and detoxification of anthraquinone dye reactive blue 4 by using aerobic bacterial granules. Water Res. 2017;122:603-613. doi:10.1016/j.watres.2017.06.005
-
Agarwal D, Dhotre D, Patil R, Shouche Y, Juvekar S, Salvi S. Potential of health and demographic surveillance system in asthma and chronic obstructive pulmonary disease microbiome research. Front public Heal. 2017;5(August):196. doi:10.3389/fpubh.2017.00196
-
Dhotre DP, Rajabal V, Sharma A, et al. Reclassification of Phycicola gilvus (Lee et al. 2008) and Leifsonia pindariensis (Reddy et al. 2008) as Microterricola gilva comb. nov. and Microterricola pindariensis comb. nov. and emended description of the genus Microterricola. Int J Syst Evol Microbiol. 2017;67(8):2766-2772. doi:10.1099/ijsem.0.002019
-
Kumbhare S V., Kumar H, Chowdhury SP, et al. A cross-sectional comparative study of gut bacterial community of Indian and Finnish children. Sci Rep. 2017;7(1):10555. doi:10.1038/s41598-017-11215-y
-
Ghosh B, Gaike A, Kumbhare S, et al. Induced sputum microbiome in smoker and non-smoker COPD subjects and its association with lung function in Indian subjects. 2017:OA4412. doi:10.1183/1393003.congress-2017.oa4412
-
Shetty SA, Bodkhe R, Dhotre DP, et al. Comparison of small gut and whole gut microbiota of first-degree relatives with adult patients with celiac disease and controls. bioRxiv. 2017:227272.
-
Joshi A, Thite S, Kulkarni G, et al. Nitrincola alkalisediminis sp. nov., an alkaliphilic bacterium isolated from an alkaline lake. Int J Syst Evol Microbiol. 2016;66(3):1254-1259. doi:10.1099/ijsem.0.000868
-
Bhute S, Pande P, Shetty SA, et al. Molecular characterization and meta-analysis of gut microbial communities illustrate enrichment of Prevotella and Megasphaera in Indian Subjects . Front Microbiol . 2016;7(May):1-14. doi:10.3389/fmicb.2016.00660
-
Bagade AV V, Paul D, Rikame T, et al. Diversity of arsenic resistant bacteria from Lonar lake: A meteorite impact alkaline crater lake in India. In: Arsenic Research and Global Sustainability: Proceedings of the Sixth International Congress on Arsenic in the Environment (As2016), June 19-23, 2016, Stockholm, Sweden. CRC Press; 2016:113.
-
Shaligram S, Kumbhare S V, Dhotre DP, et al. Erratum to: Genomic and functional features of the biosurfactant producing Bacillus sp. AM13. Funct Integr Genomics. 2016;17(5):557-566.
-
Agarwal D, Dhotre D, Brashier B, Shouche Y, Juvekar S, Salvi S. Oral and nasal microbiota in healthy, tobacco smoke COPD (TSCOPD) and biomass smoke COPD (BMSCOPD) subjects from India. 2016:PA3403. doi:10.1183/13993003.congress-2016.pa3403
-
Shouche YS, Dighe AS, Dhotre DP, Patole MS, Ranade DR. Selenomonas. Bergey’s Man Syst Archaea Bact. 2015:1-12.
-
Salunkhe RC, Dhotre DP, Salunke BK, et al. Distribution and molecular characterization of Wolbachia endosymbionts in odonata (insecta) from Central India by multigene approach. Curr Sci. 2015;108(5):971-978.
-
Kumbhare S V, Dhotre DP, Dhar SK, et al. Insights into Diversity and Imputed Metabolic Potential of Bacterial Communities in the Continental Shelf of Agatti Island. PLoS One. 2015;10(6):e0129864. doi:10.1371/journal.pone.0129864
-
Walujkar SA, Dhotre DP, Marathe NP, Lawate PS, Bharadwaj RS, Shouche YS. Characterization of bacterial community shift in human Ulcerative Colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut Pathog. 2014;6(1):22.
-
Mahale KN, Paranjape PS, Marathe NP, et al. Draft genome sequences of Yersinia pestis strains from the 1994 plague epidemic of Surat and 2002 Shimla outbreak in India. Indian J Microbiol. 2014;54(4):480-482. doi:10.1007/s12088-014-0475-7
-
Kulkarni G, Dhotre D, Dharne M, et al. Draft genome of Ochrobactrum intermedium strain M86 isolated from non-ulcer dyspeptic individual from India. Gut Pathog. 2013;5(1):7.
-
Gokhale K, Patil DPDP, Dhotre DPDP, et al. Transcriptome analysis of Anopheles stephensi embryo using expressed sequence tags. J Biosci. 2013;38(2):301-309. doi:10.1007/s12038-013-9320-0
-
Shetty SA, Marathe NP, Munot H, et al. Draft genome sequence of Methylophaga lonarensis MPLT, a haloalkaliphilic (non-methane-utilizing) methylotroph. Genome Announc. 2013;1(3):e00202-13.
-
Shetty S, Marathe N, Dhotre D, et al. Differences in fecal microbial community composition in celiac disease before and after gluten free diet. J Gastroenterol Hepatol. 2013;28:595-596.
-
Salunke BKBK, Salunkhe RCRC, Dhotre DPDP, et al. Determination of Wolbachia diversity in butterflies from Western Ghats, India, by a multigene approach. Appl Environ Microbiol. 2012;78(12):4458-4467. doi:10.1128/AEM.07298-11
-
Patil D, Dhotre DP, chavan S, et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J Biosci. 2012;37(4):647-657. doi:10.1007/s12038-012-9244-0
-
Pawar SP, Dhotre DP, Shetty SA, Chowdhury SP, Chaudhari BL, Shouche YS. Genome sequence of Janibacter hoylei MTCC8307, isolated from the stratospheric air. J Bacteriol. 2012;194(23):6629-6630.
-
Salunke BK, Salunkhe RC, Dhotre DP, et al. Diversity of Wolbachia in Odontotermes spp. (Termitidae) and Coptotermes heimi (Rhinotermitidae) using the multigene approach. FEMS Microbiol Lett. 2010;307(1):55-64. doi:10.1111/j.1574-6968.2010.01960.x
-
Patil DP, Atanur S, Dhotre DP, et al. Generation, annotation, and analysis of ESTs from midgut tissue of adult female Anopheles stephensi mosquitoes. BMC Genomics. 2009;10(386):386. doi:10.1186/1471-2164-10-386
-
Vaishampayan PA, Dhotre DP, Gupta RP, et al. Molecular evidence and phylogenetic afiliations of Wolbachia in cockroaches. Mol Phylogenet Evol. 2007;44:1346-1351.
-
Pandey PK, Dhotre DP, Dharne MS, et al. Evaluation of mitochondrial 12S rRNA gene in the identification of Panthera pardus fusca (Meyer, 1794) from field-collected scat samples in the Western Ghats, Maharashtra, India. Curr Sci. 2007;92(8).
Awards/Honour/Memberships
Member of Bergey’s
Awarded a DBT-JRF in the subject of Bioinformatics in July 2005.
Awarded a DBT-SRF in the subject of Bioinformatics in 2007.
Travel award for Management and application of Microbial Data Resource supported by WFCC, UNESCO, WDCM and IMCAS during 2014
Funding
Our laboratory is funded by Department of Biotechnology, Department of Science and Technology and Science and Engineering Research Board, New Delhi.
Current Lab members
Varun Shah, Ph.D (Project Scientist C)
Abhijit Kulkarni (PhD Student)
Kamala Sape (PhD Student)
Akshay Gaike (PhD Student)
Mitali Inamdar (ICMR SRF)
Ashwini Hagir (JRF)
Namrata Jiya (JRF)
Aditi Deshpande (JRF)
Shivang Bhanushali (JRF)
Swapnil Bodkhe (JRF)
Avinash Kshirsagar (JRF)
Gayatri Chandan (Project Trainee)
Bhagyashree Karmarkar (PhD Student)
Puja Ghosh (PhD Student)
Past Lab members
Abhishek Gupta, Ph.D (Research Associate III)
Mohak Gujare (JRF)
Harshada Ghode (JRF)
Opportunities
Highly motivated scientists interested in Human microbiome research with strong background in NGS, data analysis, metabolomics, anaerobic isolation and characterization and animal studies are always encouraged to apply for post-doc positions.
Students interested in MS/M.Sc project training projects (6-12 months), please email dhiraj.dhotre@nccs.res.in or academics@nccs.res.in
Back
Last updated On : 03 August 2022 09:56


